Who is here? 1 guest(s)
|
Small acalyptrate fly - Diastata flavicosta (or fuscula)
|
|
| djo |
Posted on 16-08-2023 19:37
|
|
Member Location: Posts: 187 Joined: 16.05.11 |
Aspirated from a fallen deciduous tree trunk in Sussex, UK, August 2023. The scale intervals are 0.5mm
Edited by djo on 17-08-2023 12:04 |
|
|
|
| djo |
Posted on 16-08-2023 19:38
|
|
Member Location: Posts: 187 Joined: 16.05.11 |
Aspirated from a fallen deciduous tree trunk in Sussex, UK, August 2023. The scale intervals are 0.5mm |
|
|
|
| Tony Irwin |
Posted on 17-08-2023 08:03
|
|
Member Location: Posts: 7168 Joined: 19.11.04 |
Diastata?
Tony ---------- Tony Irwin |
|
|
|
| djo |
Posted on 17-08-2023 09:22
|
|
Member Location: Posts: 187 Joined: 16.05.11 |
Hmmm .... someone else (iNat) suggested Trixoscelis (Heleomyzidae). The spikes on the costa look right for that, but the setae on the mesonotum look wrong. Diastata (cf costata?) looks like a good idea! Edited by djo on 17-08-2023 09:26 |
|
|
|
| Jan Maca |
Posted on 17-08-2023 10:11
|
|
Member Location: Posts: 1113 Joined: 25.03.10 |
Diastata, looks like D. flavicosta but a rather detailed study (arista, genitalia) is needed to distinguish it from D. fuscula. I suggest to send the specimen to Peter Chandler. |
|
|
|
| djo |
Posted on 17-08-2023 14:14
|
|
Member Location: Posts: 187 Joined: 16.05.11 |
Hmmm, looks like fuscula but not flavicosta is in the UK list. Unfortunately, I want to sequence the genome - which is not compatible with ding anything else to the specimen! |
|
|
|
| Tony Irwin |
Posted on 17-08-2023 20:27
|
|
Member Location: Posts: 7168 Joined: 19.11.04 |
Darren - you shouldn't need the whole specimen for DNA investigation. Remove the posterior half of the abdomen - this has all you need to identify the specimen morphologically (male genitalia). And the remainder of the specimen will be plenty to extract your DNA samples.
Tony ---------- Tony Irwin |
|
|
|
| djo |
Posted on 18-08-2023 09:33
|
|
Member Location: Posts: 187 Joined: 16.05.11 |
Hi! Unfortunately genome sequencing requires a lot more DNA than for PCR/Sanger for COI etc (which need <1ng DNA). Minimum for Novaseq/short-read genome might be ~20ng at ~2ng/ul, and in fact we often struggle to get enough at sufficient concentration from single Drosophila. But this would actually be going for ONT long read, which really would benefit from the whole fly. For comparison, Darwin tree of life ask for 4 specimens when doing stuff this small (Hifi,HiC,RNA, and spare!) It's part of a Drosophilidae project, but the world is desperately short of suitable outgroup genomes (i.e. Ephydroidea) and so my collaborator is searching for (fresh, -20C in Ethanol) specimens to sequence! The species really don't matter, so long as they sit between Opomyzoidea/Carnoidea and Drosophilidae! I'll check with my collaborator, and see what he thinks. Thanks! D |
|
|
|
| Jump to Forum: |












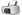
 but don't see the image in the post.
but don't see the image in the post.
